PanCanSurvPlot
PanCanSurvPlot is a large-scale interactive web tool dedicated to pan-cancer transcriptome survival analysis and visualization. With PanCanSurvPlot, users can quickly explore the impact of target genes on survival outcomes in different tumors, assisting clinicians and researchers in further investigating the mechanism of tumor development and improving clinical decision-making.
PanCanSurvPlot is accessible at: http://www.PanCanSurvPlot.com/(New) and https://smuonco.shinyapps.io/PanCanSurvPlot/.
The preprint is now online.
Citation: Lin, A., Yang, H., Shi, Y., Cheng, Q., Liu, Z., Zhang, J., & Luo, P. (2022). PanCanSurvPlot: A Large-scale Pan-cancer Survival Analysis Web Application. BioRxiv, 2022.12.25.521884. https://doi.org/10.1101/2022.12.25.521884.
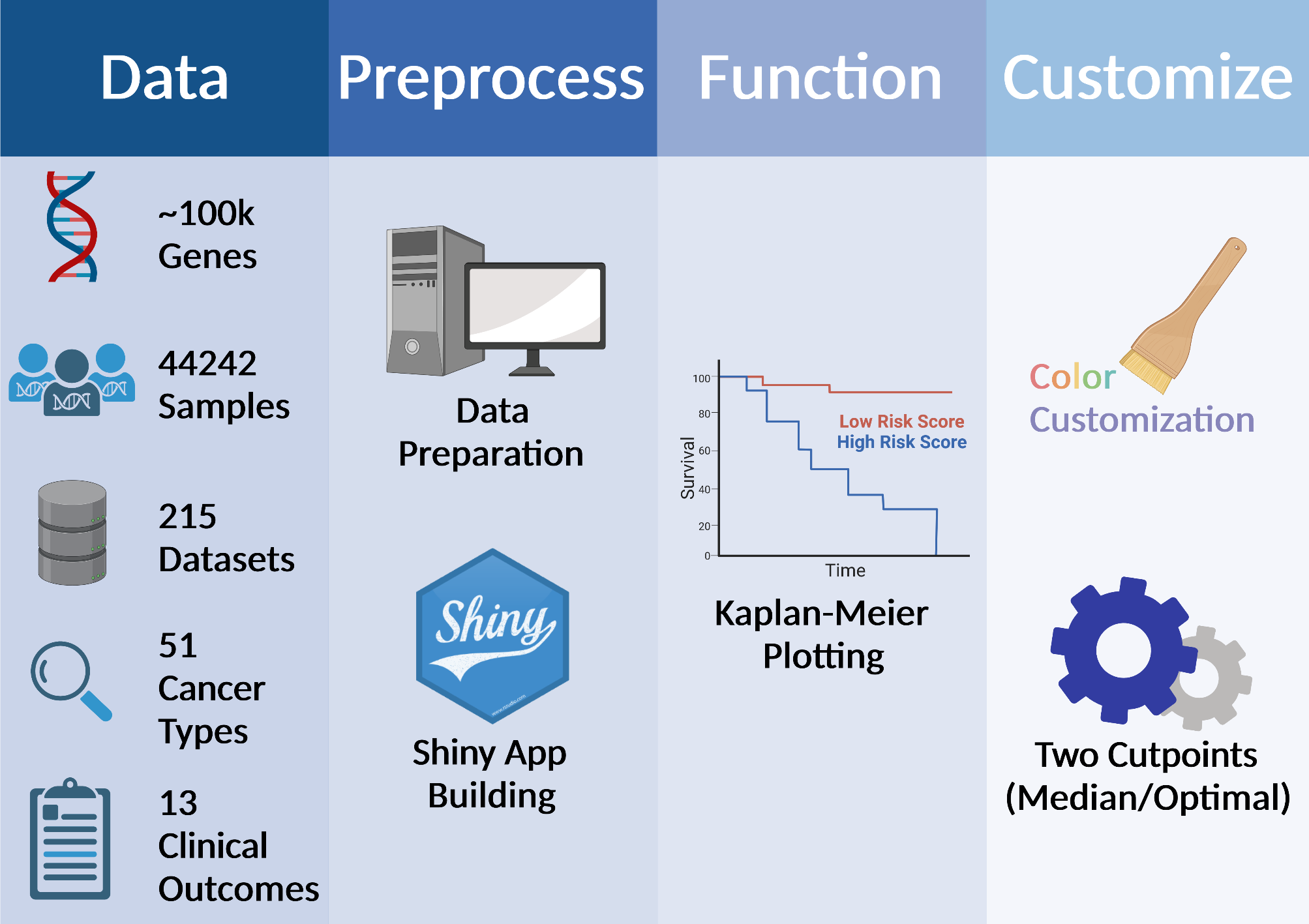
How does PanCanSurvPlot work?
STEP1: Select Data to Be Processed
STEP2: View Analysis Results
STEP3: Costomize and Download Your Plot
Tutorial
Learn more about PanCanSurvPlot on the Youtube tutorial video. For users who can not access YouTube, the tutorial video is also available on Bilibili.
Updates
19/01/23 Version 1.2.1 released. Cox results (continuous) are now available.
29/12/22 Version 1.2.0 released. Therapy Details are now available.
25/11/22 Version 1.1.0 released. TCGA data are now available.
23/10/22 Version 1.0.0 released.
STEP1: Select Data to Be Processed
STEP2: View Analysis Results
STEP3: View, Costomize and Download Your Plot
DATASETS DESCRIPTION
Contact
Should you have any questions, please feel free to contact us.
Peng Luo: luopeng@smu.edu.cn
Hong Yang: smuyanghong@i.smu.edu.cn
Ying Shi: shoshanashi@i.smu.edu.cn
Our lab has a long-standing interest in cancer biomedical research and bioinformatics. We recently developed several other Shiny web tools focusing on solving various scientific questions.
CAMOIP: A Web Server for Comprehensive Analysis on Multi-omics of Immunotherapy in Pan-cancer. doi: 10.1093/bib/bbac129
Onlinemeta: A Web Server For Meta-Analysis Based On R-shiny. doi: 10.1101/2022.04.13.488126[preprint]
Tutorial Video
For users who can not access YouTube, the tutorial video is also available on Bilibili.
Comment Box
Update History
19/01/23 Version 1.2.1 of PanCanSurvPlot released.
NEW FEATURE: Cox results (continuous) have been added to the results summary table.
29/12/22 Version 1.2.0 of PanCanSurvPlot released.
NEW FEATURE: Therapy Details are now available. The preprint is now online.
25/11/22 Version 1.1.0 of PanCanSurvPlot released.
NEW FEATURE: TCGA data are now available. Bugs of sorting the scientific notation were repaired.
23/10/22 Version 1.0.0 of PanCanSurvPlot released.
08/10/22 Version 1.0.0beta of PanCanSurvPlot released.
FAQ
1. Why does the gene symbol I type in return no results?
The gene symbols we provided here mostly come from HGNC-approved protein-coding genes, noncoding RNA genes and pseudogenes. The whole list of genes we provided can be found here.
2. What are the full forms of the survival outcomes provided?
Altogether, there are 13 different survival outcomes available. Their abbreviations and corresponding full forms are shown below.
BCR: Biochemical Recurrence Free Survival
CSS: Cancer Specific Survival
DFI: Disease Free Interval
DFS: Disease Free Survival
DMFS: Distant Metastasis Free Survival
DRFS: Distant Relapse Free Survival
DSS: Disease Specific Survival
FFS: Failure Free Survival
MFS: Metastasis Free Survival
OS: Overall Survival
PFI: Progression Free Interval
PFS: Progression Free Survival
RFS: Recurrence Free Survival
3. Why is there no median survival line shown in the K-M plot I have made?
Because of the specific cutoff point you chose, the median survival has not yet been reached, and more than half of the patients are still alive.
4. What is the difference between COX results (continuous) and COX results with the median/best cutpoint? How are they calculated?
For COX results (continuous), we treated the patient survival data as a continuous variable and fitted a Cox proportional hazards regression model to them. For COX results with the median/best cutpoint, we grouped the patient survival data according to the median/best cutpoint and performed the bivariate Cox proportional hazards regression.
5. Why does the p-value and confidence interval of the hazard ratio disagree, with the p-value < 0.05 but the 95% confidence interval of the hazard ratio covers 1.0 or vice versa?
Due to reasons such as small sample sizes or uneven groupings in some of the datasets, the high level of sampling error might cause the inconsistency between p value and 95% confidence interval.